We had a go at a simple, new algorithm based on Bradford cone space. Yoav Aner from kenhub has made a plugin for carrierwave based on it, and there is also a command-line version and an implementation in nip2.
Background
Daltonization is the process of adjusting the colour of images to make them easier for colour-blind people to interpret. For example, consider this photograph from the vischeck website:A person with Deuteranopia, deficient green receptors, might see something like this:
How many types of fruit are there? It's rather hard to tell. If we remap the missing red-green information to the other colour channels we could make something like this:
And now, presented with this image, a Deutanerope might see:
The fruit which looked the same now looks different.
Simulating colour-blindness
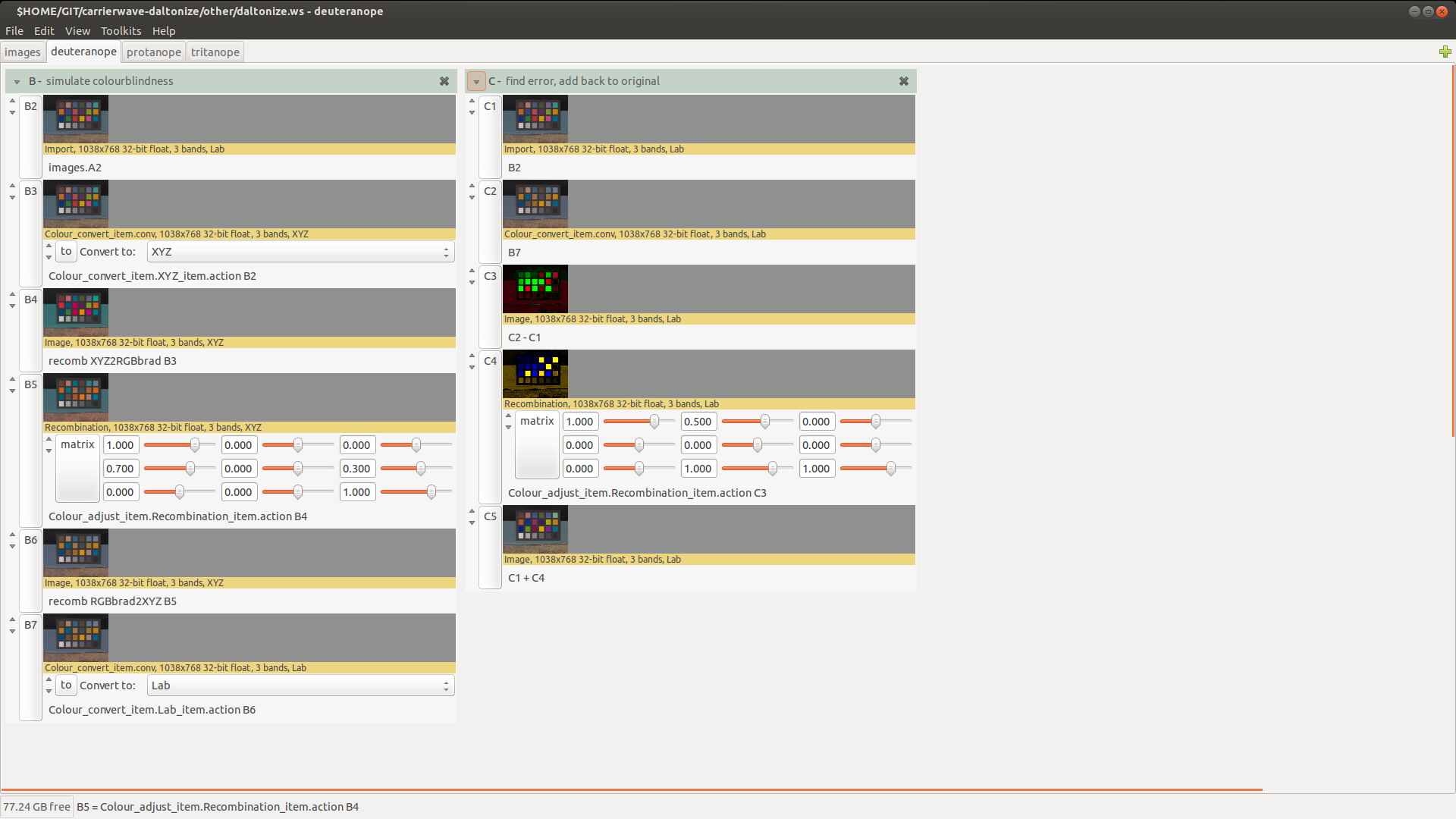
The first step is to be able to simulate colour-blindness. We do this by importing the RGB image to CIE XYZ using the image's attached profile, then transforming to Bradford cone space. In this colourspace the three channels correspond roughly to the three cone responses.For Deutaneropia, we need to ignore the green channel. We could just zero the middle channel, but that would make an image which looked horrible to non-colour-blind people, so instead we replace it with a 70% to 30% mix of the red and blue channels. This will throw away the green information while leaving the neutral colours neutral. We choose a 70:30 mix, since red receptors are much more common than blue. Since we are recombining colour channels, we remove the D65 cast that XYZ will have.
We then go back to CIE XYZ, and this gives us the simulated Deutanerope vision image above.
In Ruby-vips, we can write this as:
# D65 to E and back
D652E = Matrix.diagonal(100 / 95.047, 100 / 100, 100 / 108.883)
E2D65 = D652E ** -1
# Take CIE XYZ to Bradford cone space (a variant of LMS)
XYZ2BRAD = Matrix[[0.8951, 0.2664, -0.1614],
[-0.7502, 1.7135, 0.0367],
[0.0389, -0.0685, 1.0296]]
BRAD2XYZ = XYZ2BRAD ** -1
begin
# import to CIELAB with lcms
# if there's no profile there, we'll fall back to the thing below
cielab = image.icc_import_embedded(:relative)
xyz = cielab.lab_to_xyz()
rescue VIPS::Error
# nope .. use the built-in srgb converter instead
xyz = image.srgb_to_xyz()
cielab = xyz.xyz_to_lab()
end
# simulate deutaneropia
simulate = [[ 1, 0, 0],
[0.7, 0, 0.3],
[ 0, 0, 1]]
# pre-multiply our colour matrix
m = E2D65 * BRAD2XYZ * Matrix.rows(simulate) * XYZ2BRAD * D652E
# simulate colour-blindness
xyz2 = xyz.recomb(m.to_a())
cielab2 = xyz2.xyz_to_lab()
Remapping the missing information
Now we can just subtract the simulated colour-blind image from the original to get an approximation of the missing information. CIE LAB is a convenient space to do this. Then we need to remap the missing information to channels which are perceptible to the colour-blind person, and add that to the original image.There's some choice about the best way to do this. A deutanerope will be deficient in red-green, so we decided to add 100% of the error in the a* channel to the yellow-blue b* channel, and 50% to the L* lightness channel.
In Ruby-vips this would be:
# find the colour errorAnd that produces the compensated image above.
err = cielab - cielab2
# add the error channels back to the original, recombined so as to hit
# channels the person is sensitive to
cielab = cielab + err.recomb([[ 1, 0.5, 0],
[ 0, 0, 0],
[ 0, 1, 1]])
# .. and back to sRGB
rgb = cielab.lab_to_xyz().xyz_to_srgb()
Protaniopia and Tritanopia, deficiencies in the red and the blue receptors respectively, need different matrices for simulation and error distribution. We've used these in our algorithm:
protanioia_simulate = [[ 0, 0.8, 0.2],
[ 0, 1, 0],
[ 0, 0, 1]]
protanopia_distribute = [[ 1, 0.5, 0],
[ 0, 0, 0],
[ 0, 1, 1]]
tritanopia_simulate = [[ 1, 0, 0],
[ 0, 1, 0],
[0.3, 0.7, 0]]
tritanopia_distribute = [[ 1, 0, 0.5],
[ 0, 0, 1],
[ 0, 0, 0]]
Comparison to other work
There are quite a few Daltonization algorithms around. We found a Python implementation of one of the most widely-used daltonization algorithms to compare our work to.We made a test image showing a set of Ishihara blobs and the same set processed by the usual algorithm and by our modified one:
We put this test image online and asked colour-blind people on several web forums to rank the left, middle and right sets by legibility. We didn't say which was which.
Our algorithm (the centre column) won convincingly (fortunately for us).
Speed
On my laptop the filter runs reasonably quickly. For a 1,000 x 1,000 pixel RGB JPEG I see:$ time ./daltonize.rb ~/pics/Gugg_coloured.jpg ~/x.jpg deuteranope
real 0m0.270s
user 0m0.551s
sys 0m0.040s
The Python filter is slow for some reason, I'm sure it could be sped up. On the same image I see:
$ time ../daltonize.py ~/pics/Gugg_coloured.jpg
Daltonize image correction for color blind users
Please wait. Daltonizing in progress...
Creating Deuteranope corrected version
Image successfully daltonized
real 0m24.623s
user 0m23.256s
sys 0m0.128s
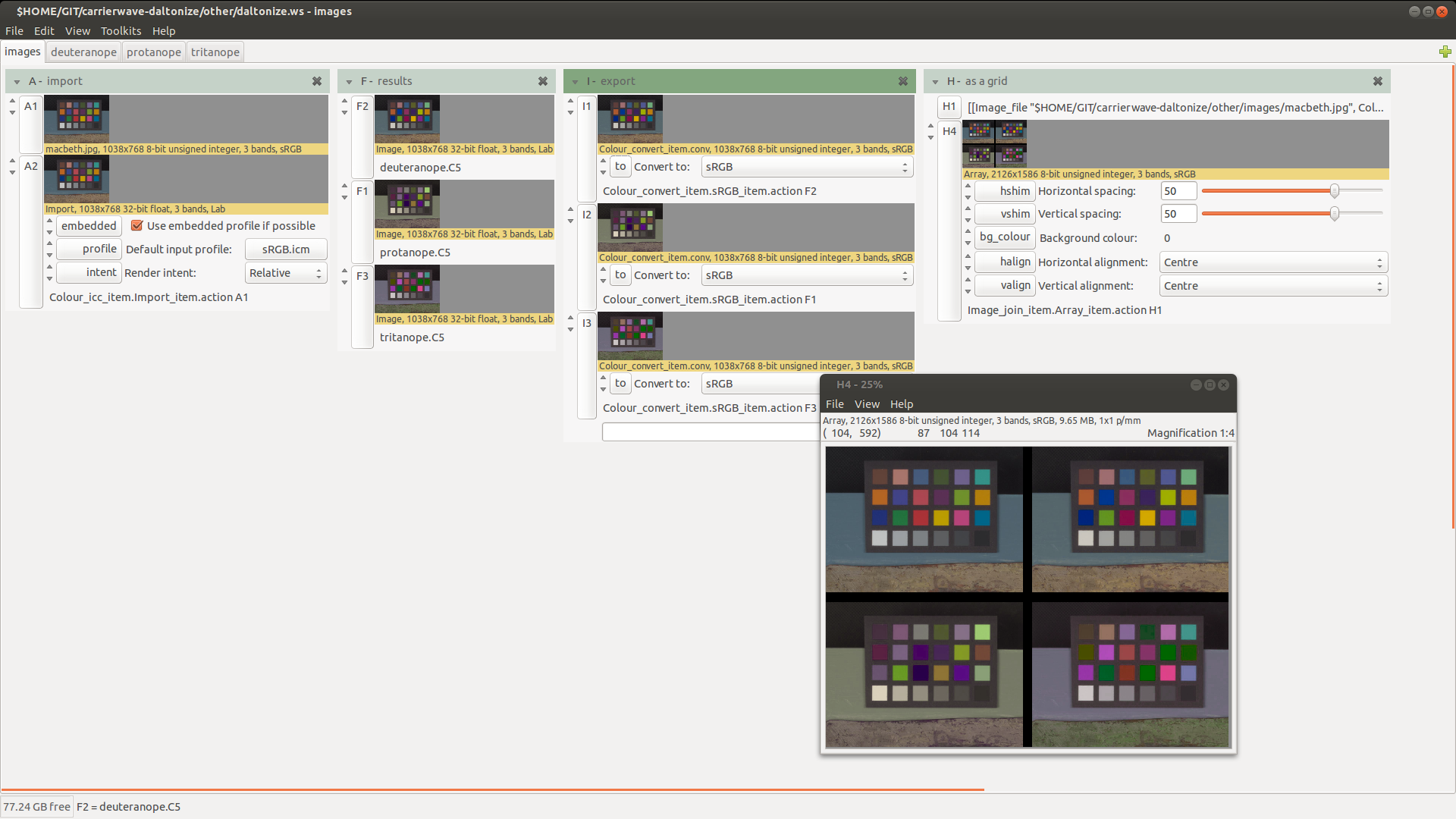
As a nip2 workspace
We also made a nip2 workspace implementing this algorithm. You'll need nip2 7.33 or later to try this out.It's quite neat: you can drag in an image and it'll make you images tweaked for the three major typs of colour-blindness.
You can switch to the tab for a certain vision type and edit the matrices to see the effect of changes:






No comments:
Post a Comment